Reconstructing the evolutionary history of a remarkable moss family.
Significance of autopolyploidy in Physcomitrium pyriforme
Website of the Funariaceae Research Group

NSF funded grant (DEB-1753673): Diversity of Physcomitrium pyriforme in North America and Europe: significance of autopolyploidy within a phylogenomic and experimental framework. A collaborative project with Dr. Bernard Goffinet (UConn) and Dr. Matt Johnson (Texas Tech. University)
 We are using the goblet moss (Physcomitrium pyriforme) as a model organism to learn about the role of genome duplication in plant evolution. The research we do involves culturing different strains, generation of polyploids, morphometrics and experiments on reproductive isolation.
We are using the goblet moss (Physcomitrium pyriforme) as a model organism to learn about the role of genome duplication in plant evolution. The research we do involves culturing different strains, generation of polyploids, morphometrics and experiments on reproductive isolation.
This project is also developing a citizen science component that we call Physco Hunt. Anybody in North America or Europe can contribute finding, reporting and sending specimens for us to culture and do our research. Check the project on iNaturalist where we monitor recent finds of this species. For those who are willing to find this moss around them, download the training document.
If you want to help us with this project, join the PhyscoHunt! You can report and send the samples of Physcomitrium that you find anywhere in North America or Europe. Download the guidelines for collaboration here.
 Plants are weird. Many of the mechanisms that result in the birth and diversification of new plant species differ from those that have been discovered using animal models, which are the majority. One of the favorite tricks of plant evolution involves the duplication of the genome (autopolyploidy: cells gain a complete extra set of chromosomes). In fact, we know that numerous plant evolutionary bursts were correlated with autopolyploidy events, including many that gave rise to current species of economic interest. However, the evolutionary significance of autopolyploidy is still not completely understood despite its relevance. Whole genome duplications are often difficult to detect, and at best we see their outcomes long after they occurred, since they are not always easy to observe under lab conditions. In this project we will address the role and consequences of polyploidy in plant evolution using as model organism the goblet moss (Physcomitrium pyriforme).
Plants are weird. Many of the mechanisms that result in the birth and diversification of new plant species differ from those that have been discovered using animal models, which are the majority. One of the favorite tricks of plant evolution involves the duplication of the genome (autopolyploidy: cells gain a complete extra set of chromosomes). In fact, we know that numerous plant evolutionary bursts were correlated with autopolyploidy events, including many that gave rise to current species of economic interest. However, the evolutionary significance of autopolyploidy is still not completely understood despite its relevance. Whole genome duplications are often difficult to detect, and at best we see their outcomes long after they occurred, since they are not always easy to observe under lab conditions. In this project we will address the role and consequences of polyploidy in plant evolution using as model organism the goblet moss (Physcomitrium pyriforme).
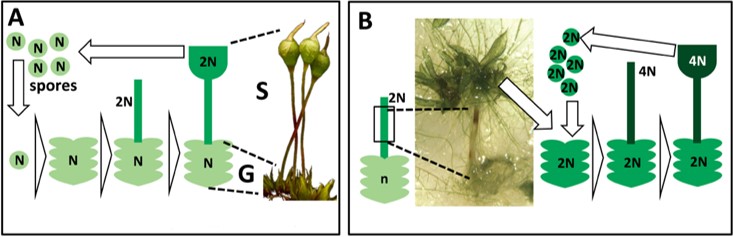
Normal life cycle of the goblet moss (A) and process to produce artificial autopolyploids in the lab (B)
This moss is ideally suited for this purpose: it harbors a morphologically-diverse complex of seven karyotypes, it is easily grown in the laboratory, and its genome content can be doubled with precision in vitro to create autopolyploid offspring. The goals of this collaborative project are to produce a phylogeny of the Physcomitrium pyriforme species-complex based on genome sequence data, to estimate past changes of genomic load over evolutionary time, and to compare reproductive barriers among naturally-occurring and artificially-induced autopolyploids.
In our lab we will be generating artificial autopolyploids, testing their fertility and their reproductive compatibility with their wild counterparts, and performing morphometric analyses to detect changes in the phenotype produced by the whole genome duplications. If you are a student in Augie and want to be a part of this, contact us!
Project overview: Whole genome duplication or autopolyploidy occurred repeatedly during the evolution of land plants and likely act as a major driver of evolutionary change. Such duplications first occurred within species, likely reproductively isolating autopolyploids. They may also trigger significant genomic restructuring, preventing meiotic pairing and hence interbreeding between two independent autodiploids. Genome duplications are thus effective mechanisms of microevolutionary change. They characterize many macroevolutionary lineages, and are expected to be frequent enough within species, for some to give rise to new evolutionary lineages. This project seeks to test whether shifts in ploidy are phylogenetically structured within a complex of cryptic moss species, the Physcomitrium pyriforme complex. It harbors seven karyotypes worldwide and exhibits much morphological variation, as reflected by its 29 synonyms. These annual, bisexual and selfing mosses are easily grown, and genome doubling is readily induced in vitro from sporophytic tissue, enabling tests of reproductive isolation among wild and artificial autopolyploids. This project addresses four objectives toward assessing the evolutionary significance of autopolyploidy: (1) reconstruct the phylogeny of the complex based on sub genome data,(2) infer frequencies of ploidal shifts within the complex; (3) identify morphological signatures of artificial genome duplication and through comparison with wild populations test whether these erode through time; and (4) test if wild and artificial polyploids are reproductively isolated.
This project is funded by the National Science Foundation (grant DEB-1753673) in collaboration with with Dr. Bernard Goffinet (UConn) and Dr. Matt Johnson (Texas Tech. University)